
Our Science
Deregulated transcription is a hallmark of cancer
Kronos Bio is focused on developing medicines that target deregulated transcription factor activity, a hallmark of cancer and other serious diseases. In response to various cell signals, transcription factors activate or repress gene expression to determine cell identity and function. Regulation of transcription factors is important for processes such as normal development, but when transcription factors are deregulated, they can drive a cell to become malignant. For decades, transcription factors have been sought after as therapeutic targets. Until now, researchers have pursued transcription factors as isolated proteins – resulting in little success.
Our approach is different. We pursue transcription factors in context.
By analyzing transcription factors as part of complexes and networks, we can identify the critical nodes in these signaling pathways that are responsible for a transcription factor’s activity. This provides multiple targets for us to pursue drug development, rather than limiting our focus to a single transcription factor.
We believe there is significant potential in this space – only a handful of the more than 100 transcription factors implicated in driving cancer have been drugged.
Our President & CEO Dr. Norbert Bischofberger explains more. In the first video, Norbert talks about the role that transcription plays in normal human activity. In the second, he talks about how the connection between dysregulated transcription factor activity and cancer.
Hear from our CEO
Join our President and CEO Norbert Bischofberger at the white board, as he explains how transcription works in normal cells (above) and in cancer cells (below).
Why is it so Important to Drug Transcription Factors in Context?
Transcription factors are proteins that bind DNA and turn genes on and off. Transcription factors in isolation are mostly unstructured, with the exception of the DNA binding domain, providing limited opportunity to identify binding pockets for therapeutics.
By looking at a transcription factor in the cellular context with its cofactors, researchers gain a much richer picture of transcription factor function and are able to access a broader druggable space. Transcription factors – and their cofactors – form complexes that work with the transcription apparatus on the DNA to turn on and off nearby target genes. These complexes, along with other signaling proteins, make up the transcription regulatory network. By analyzing transcription factors in various cellular contexts, such as cell types (e.g., lung, blood) or states (e.g., oncogenic vs. normal), we can identify whether different cofactors assemble to make unique transcription regulatory complexes to regulate genes within that specific context.
Targeting Oncogenic Transcription Regulatory Networks
At Kronos Bio, we are focused on oncogenic transcription factors, such as MYC, which have a well-defined and central role in driving particular types of cancer. We leverage our computational biology expertise to identify protein-protein interactions, genetic dependencies and gene expression signatures to better understand specific transcription factors in cancers of interest. We take this knowledge and use it to map the transcription regulatory network. This map allows us to identify known and previously unknown protein interactions and genetic dependencies. Within a transcription regulatory network, we can identify complex relationships, such as positive feedback loops, which only exist in tumors.
Our understanding of these complex relationships within a transcription regulatory network offers potential avenues to target tumors and help us identify which patients may be mostly likely to benefit from a particular approach.
When drugging known targets, we can use this knowledge to optimize the therapeutic approach. It also allows us to leverage transcription regulatory network signatures to identify new targets using our Small Molecule Microarray (SMM) to screen for new drug starting points.
Our SMM Screening Platform
We use our Small Molecule Microarray (SMM) product engine to conduct high throughput binding assays against traditionally undruggable transcription factors in tumor cell lysate. By conducting screens in cell lysate, we maintain endogenous protein structures and complexes, allowing us to probe the entire transcription factor complex in a single assay.
Hits identified in SMM lysate screens bind to the transcription factor complex and may be binders of the transcription factors or their immediate co-factors. We use transcription regulatory network signatures to prioritize hits that modulate abnormal transcription factor activity. These hits have the potential to modulate this activity through a number of possible mechanisms, including inhibition, and stabilization or destabilization of target proteins or complexes.
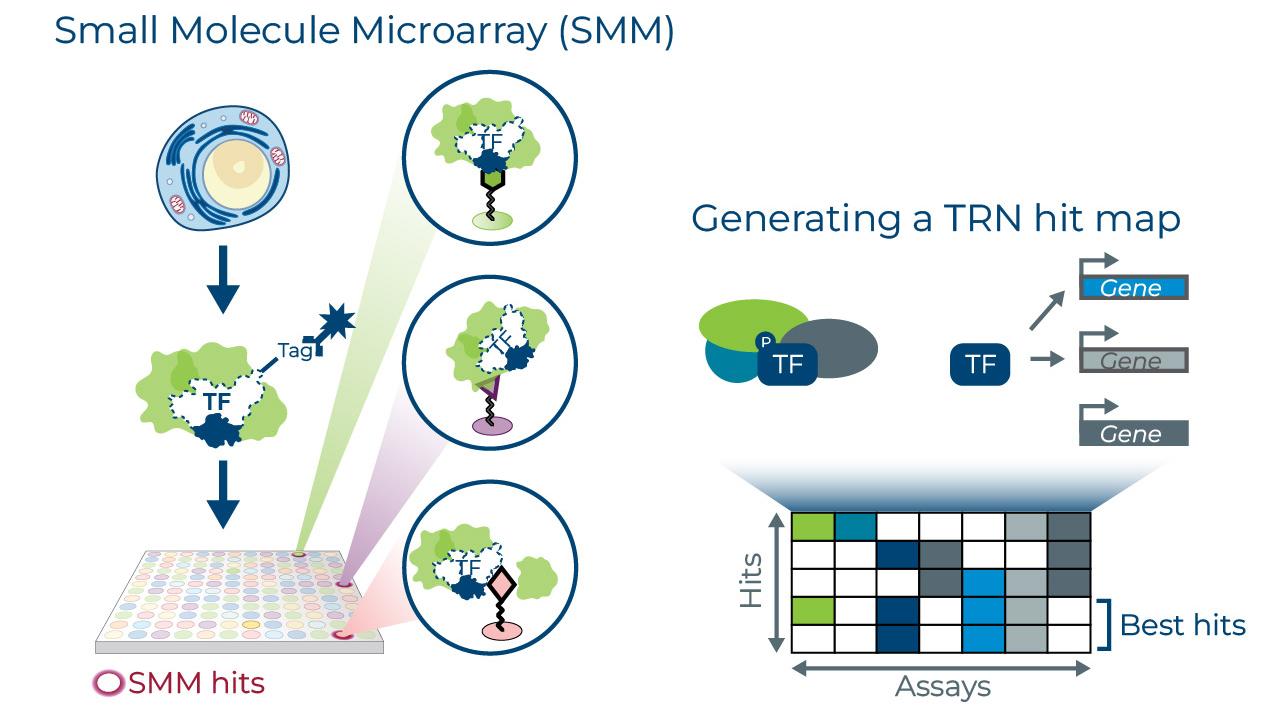
More About the SMM
- Licensed from the lab of Dr. Angela Koehler, Ph.D., at the Massachusetts Institute of Technology
- Includes 240,000 compound library covalently printed on slides
- Allows screening of cell lysates/nuclear extracts
Learn More About our Pipeline
At Kronos Bio, we have two molecules that have emerged from our product engine. KB-0742, our CDK9 inhibitor, is being evaluated in a phase 1/2 clinical trial as a treatment for patients with MYC-dependent tumors such as triple negative breast cancer, non-small cell lung cancer and ovarian cancer, as well as patients with transcription factor fusion-driven cancers and other transcriptionally addicted cancers including chordomas, sarcomas and small cell lung cancer. Our new development candidate, KB-9558, targets the KAT domain of p300, a critical node of the IRF4 TRN, which is a core oncogenic transcription program that drives multiple myeloma.
Additionally, Kronos Bio began a discovery collaboration with Genentech in January 2023. Our companies are working together to advance novel therapies against transcriptional targets in oncology.
To learn more please visit our pipeline and scientific publications pages.